Bipolar disorder genetic component
Genetics of bipolar disorder - PMC
1. Angst F., Stassen HH., Clayton PJ., Angst J. Mortality of patients with mood disorders: follow-up over 34-38 years. J Affect Disord. 2002;68:167–181. [PubMed] [Google Scholar]
2. Tondo L., Baldessarini RJ. Reduced suicide risk during lithium maintenance treatment. J Clin Psychiatry. 2000;61(suppl 9):97–104. [PubMed] [Google Scholar]
3. Namjoshi MA., Buesching DP. A review of the health-related quality of life literature in bipolar disorder. Qual Life Res. 2001;10:105–115. [PubMed] [Google Scholar]
4. Wyatt RJ., Henter I. An economic evaluation of manic-depressive illness - 1991. Soc Psychiatry Psychiatr Epidemiol. 1995;30:213–219. [PMC free article] [PubMed] [Google Scholar]
5. Begley CE., Annegers JF., Swann AC., et al The lifetime cost of bipolar disorder in the US: an estimate for new cases in 1998. Pharmacoeconomics. 2001;19:483–495. [PubMed] [Google Scholar]
6. Goodwin Kaj., KR Manic. Depressive Illness. New York, NY: Oxford University Press. 1990 [Google Scholar]
7. Escamilla MA., Reus VI., Freimer NB. Molecular and Genetic Basis of Neurological Disease. London, UK: Butterworth-Heinemann. 1997 [Google Scholar]
8. Tsuang M., Faraone S. The Genetics of Mood Disorders. Baltimore, Md: Johns Hopkins Press. 1990 [Google Scholar]
9. Gershon E., Hamovit J., Guroff JJ., et al A family study of schizoaffective, bipolar I, bipolar II, unipolar, and normal control probands. Arch Gen Psychiatry. 1982;39:1157–1167. [PubMed] [Google Scholar]
10. Craddock N., Khodel V., Van Eerdewegh P., Reich T. Mathematical limits of multilocus models: the genetic transmission of bipolar disorder. Am J Hum Genet. 1995;57:690–702. [PMC free article] [PubMed] [Google Scholar]
11. Bertelsen A., Harvald B., Hauge M. A Danish twin study of manic-depressive disorders. Br J Psychiatry. 1997;130:330–351. [PubMed] [Google Scholar]
[PubMed] [Google Scholar]
12. Merikangas KR., Chakravarti A., Moldin SO., et al Future of genetics of mood disorders research. Biol Psychiatry. 2002;52:457–477. [PubMed] [Google Scholar]
13. Craddock N., Van Eerdewegh P., Reich T. Single major locus models for bipolar disorder are implausible. Am J Med Genet. 1997;74:18–20. [PubMed] [Google Scholar]
14. Stine OC., Xu J., Koskela R., et al Evidence for linkage of bipolar disorder to chromosome 18 with a parent-of-origin effect. Am J Hum Genet. 1995;57:1384–1394. [PMC free article] [PubMed] [Google Scholar]
15. Freimer NB., Reus VI., Escamilla MA., et al Genetic mapping using haplotype, association and linkage methods suggests a locus for severe bipolar disorder (BPI) at 18q22-q23. Nat Genet. 1996;12:436–441. [PubMed] [Google Scholar]
16. Escamilla MA., Mclnnes LA., Spesny M., et al Assessing the feasibility of linkage disequilibrium methods for mapping complex traits: an initial screen for bipolar disorder loci on chromosome 18. Am J Hum Genet. 1999;64:1670–1678. [PMC free article] [PubMed] [Google Scholar]
Am J Hum Genet. 1999;64:1670–1678. [PMC free article] [PubMed] [Google Scholar]
17. McMahon FJ., Hopkins PJ., Xu J., et al Linkage of bipolar affective disorder to chromosome 18 markers in a new pedigree series. Am J Hum Genet. 1997;61:1397–1404. [PMC free article] [PubMed] [Google Scholar]
18. Blackwood DH., He L., Morris SW., et al A locus for bipolar affective disorder on chromosome 4p. Nat Genet. 1996;12:427–430. [PubMed] [Google Scholar]
19. Ginns El., St Jean P., Philibert RA., et al A genome-wide search for chromosomal loci linked to mental health wellness in relatives at high risk for bipolar affective disorder among the Old Order Amish. Proc Natl Acad Sel U S A. 1998;95:15531–15536. [PMC free article] [PubMed] [Google Scholar]
20. Detera-Wadleigh SD., Badner JA., Berrettini WH., et al A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11. 2. Proc Natl Acad Sci U S A. 1999;96:5604–5609. [PMC free article] [PubMed] [Google Scholar]
2. Proc Natl Acad Sci U S A. 1999;96:5604–5609. [PMC free article] [PubMed] [Google Scholar]
21. Kelsoe JR., Spence MA., Loetscher E., et al A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci U SA. 2001;98:585–590. [PMC free article] [PubMed] [Google Scholar]
22. Morissette J., Villeneuve A., Bordeleau L., et al Genome-wide search for linkage of bipolar affective disorders in a very large pedigree derived from a homogeneous population in quebec points to a locus of major effect on chromosome 12q23-q24. Am J Med Genet. 1999;88:567–587. [PubMed] [Google Scholar]
23. Moldin SO. NIMH Human Genetics Initiative: 2003 update. Am J Psychiatry. 2003;160:621–622. [PubMed] [Google Scholar]
24. Straub RE., Lehner T., Luo Y., et al A possible vulnerability locus for bipolar affective disorder on chromosome 21q22.3. Nat Genet. 1994;8:291–296. [PubMed] [Google Scholar]
25. Pekkarinen P., Terwilliger J., Bredbacka PE., Lonnqvist J., Peltonen L. Evidence of a predisposing locus to bipolar disorder on Xq24-q27.1 in an extended Finnish pedigree. Genome Res. 1995;5:105–115. [PubMed] [Google Scholar]
Pekkarinen P., Terwilliger J., Bredbacka PE., Lonnqvist J., Peltonen L. Evidence of a predisposing locus to bipolar disorder on Xq24-q27.1 in an extended Finnish pedigree. Genome Res. 1995;5:105–115. [PubMed] [Google Scholar]
26. Berrettini WH. Molecular linkage studies of bipolar disorders. Bipolar Disord. 2001;3:276–283. [PubMed] [Google Scholar]
27. Mclnnes LA., Escamilla MA., Service SK., et al A complete genome screen for genes predisposing to severe bipolar disorder in two Costa Rican pedigrees. Proc Natl Acad Sci U S A. 1996;93:13060–13065. [PMC free article] [PubMed] [Google Scholar]
28. Ophoff RA., Escamilla MA., Service SK., et al Genomewide linkage disequilibrium mapping of severe bipolar disorder in a population isolate. Am J Hum Genet. 2002;71:565–574. [PMC free article] [PubMed] [Google Scholar]
29. Liu J., Juo SH., Dewan A., et al Evidence for a putative bipolar disorder locus on 2p13-16and other potential loci on 4q31:7q34:8q13:9q31:10q2124:13q32:14q21 and 17q11-12. Mol Psychiatry. 2003;8:333–342. [PubMed] [Google Scholar]
Mol Psychiatry. 2003;8:333–342. [PubMed] [Google Scholar]
30. Herzberg I., Jasinska A., Garcia J., et al Convergent linkage evidence from two Latin-American population isolates supports the presence of a susceptibility locus for bipolar disorder in 5q31-34. Hum Mol Genet. 2006;15:3146–3153. [PubMed] [Google Scholar]
31. Kerner B., Brugman DL., Freimer NB. Evidence of linkage to psychosis on chromosome 5q33-34 in pedigrees ascertained for bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2007;144:74–78. [PubMed] [Google Scholar]
32. McQueen MB., Devlin B., Faraone SV., et al Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility loci on chromosomes 6q and 8q. Am J Hum Genet. 2005;77:582–595. [PMC free article] [PubMed] [Google Scholar]
33. Ginns El., Ott J., Egeland JA., et al A genome-wide search for chromosomal loci linked to bipolar affective disorder in the Old Order Amish. Nat Genet. 1996;12:431–435. [PubMed] [Google Scholar]
Nat Genet. 1996;12:431–435. [PubMed] [Google Scholar]
34. Pato CIM., Middleton FA., Gentile KL., et al Genetic linkage of bipolar disorder to chromosome 6q22 is a consistent finding in Portuguese subpopulations and may generalize to broader populations. Am J Med Genet B Neuropsychiatr Genet. 2005;134:119–121. [PubMed] [Google Scholar]
35. Segurado R., Detera-Wadleigh SD., Levinson DF., et al Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: Bipolar disorder. Am J Hum Genet. 2003;73:49–62. [PMC free article] [PubMed] [Google Scholar]
36. Dick DM., Foroud T., Flury L., et al Genome-wide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am J Hum Genet. 2003;73:107–114. [PMC free article] [PubMed] [Google Scholar]
37. Cassidy F., Zhao C., Badger J., et al Genome-wide scan of bipolar disorder and investigation of population stratification effects on linkage: support for susceptibility loci at 4q21:7q36:9p21:12q24:14q24, and 16p13. Am J Med Genet B Neuropsychiatr Genet. 2007;144:791–801. [PubMed] [Google Scholar]
Am J Med Genet B Neuropsychiatr Genet. 2007;144:791–801. [PubMed] [Google Scholar]
38. Stine OC., McMahon FJ., Chen L., et al Initial genome screen for bipolar disorder in the NIMH genetics initiative pedigrees: chromosomes 2:11:13:14, and X. Am J Med Genet. 1997;74:263–269. [PubMed] [Google Scholar]
39. Vazza G., Bertolin C., Scudellaro E., et al Genome-wide scan supports the existence of a susceptibility locus for schizophrenia and bipolar disorder on chromosome 15q26. Mol Psychiatry. 2007;12:87–93. [PubMed] [Google Scholar]
40. Pekkarinen P., Terwilliger J., Bredbacka PE., Lônnqvist J., Peltonen L. Evidence of a predisposing locus to bipolar disorder on Xq24-q27.1 in an extended Finnish pedigree. Genome Res. 1995;5:105–115. [PubMed] [Google Scholar]
41. Zandi PP., Willour VL., Huo Y., et al Genome scan of a second wave of NIMH genetics initiative bipolar pedigrees: chromosomes 2:11:13:14, and X. Ami J Med Genet B Neuropsychiatr Genet. 2003;119:69–76. [PubMed] [Google Scholar]
2003;119:69–76. [PubMed] [Google Scholar]
42. Pulver AE., Lasseter VK., Kasch L., et al Schizophrenia: a genome scan targets chromosomes 3p and 8p as potential sites of susceptibility genes. Am J Med Genet. 1995;60:252–260. [PubMed] [Google Scholar]
43. Blouin JL., Dombroski BA., Nath SK., et al Schizophrenia susceptibility loci on chromosomes 13q32 and 8 p21. Nat Genet. 1998;20:70–73. [PubMed] [Google Scholar]
44. Stefansson H., Sigurdsson E., Steinthorsdottir V., et al Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. [PMC free article] [PubMed] [Google Scholar]
45. Chumakov I., Blumenfeld M., Guerassimenko O., et al Genetic and physiological data implicating the new human gene G72 and the gene for Daminoacid oxidase in schizophrenia. Proc Natl Acad Sci U S A. 2002;99:13675–13680. [PMC free article] [PubMed] [Google Scholar]
46. Cloninger CR. The discovery of susceptibility genes for mental disorders. Proc Natl Acad Sci U S A. 2002;99:13365–13367. [PMC free article] [PubMed] [Google Scholar]
Proc Natl Acad Sci U S A. 2002;99:13365–13367. [PMC free article] [PubMed] [Google Scholar]
47. Lambert D., Middle F., Hamshere ML., et al Stage 2 of the Wellcome Trust UK-Irish bipolar affective disorder sibling-pair genome screen: evidence for linkage on chromosomes 6q16-q21:4q12-q21:9p21:10p14-p12 and 18q22. Mol Psychiatry. 2005;10:831–841. [PubMed] [Google Scholar]
48. McMahon FJ., Simpson SG., Mclnnis MG., Badner JA., MacKinnon DF., DePaulo JR. Linkage of bipolar disorder to chromosome 18q and the validity of bipolar II disorder. Arch Gen Psychiatry. 2001;58:1025–1031. [PubMed] [Google Scholar]
49. Heun R., Maier W. The distinction of bipolar II disorder from bipolar I and recurrent unipolar depression: results of a controlled family study. Acta Psychiatr Scand. 1993;87:279–284. [PubMed] [Google Scholar]
50. Endicott J., Nee J., Andreasen N., Clayton P., Keller M., Coryell W. Bipolar II. Combine or keep separate? J Affect Disord. 1985;8:17–28. [PubMed] [Google Scholar]
1985;8:17–28. [PubMed] [Google Scholar]
51. Nurnberger Jl Jr., Blehar MC., Kaufmann CA., et al Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–859. [PubMed] [Google Scholar]
52. Simpson SG., McMahon FJ., Mclnnis MG., et al Diagnostic reliability of bipolar II disorder. Arch Gen Psychiatry. 2002;59:736–740. [PubMed] [Google Scholar]
53. Rice JP., McDonald-Scott P., Endicott J., et al The stability of diagnosis with an application to bipolar II disorder. Psychiatry Res. 1986;19:285–296. [PubMed] [Google Scholar]
54. Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. [PubMed] [Google Scholar]
55. Cloninger CR., Kaufmann CA., Faraone SV., et al Genome-wide search for schizophrenia susceptibility loci: the NIMH Genetics Initiative and Millennium Consortium. Am J Med Genet. 1998;81:275–281. [PubMed] [Google Scholar]
1998;81:275–281. [PubMed] [Google Scholar]
56. Egeland JA., Gerhard DS., Pauls DL., et al Bipolar affective disorders linked to DNA markers on chromosome 11. Nature. 1987;325:783–787. [PubMed] [Google Scholar]
57. Schulze T., McMahon F. Genetic linkage and association studies in bipolar affective disorder: a time for optimism. Amer J Med Genet Part C Semin Med Genet. 2003;123C:36–47. [PubMed] [Google Scholar]
58. Hamshere ML., Bennett P., Williams N., et al Genomewide linkage scan in schizoaffective disorder: significant evidence for linkage at 1q42 close to DISC1, and suggestive evidence at 22q11 and 19p13. . Arch Gen Psychiatry. 2005;62:1081–1088. [PubMed] [Google Scholar]
59. Schumacher J., Kaneva R., Jamra RA., et al Genomewide scan and finemapping linkage studies in four European samples with bipolar affective disorder suggest a new susceptibility locus on chromosome 1p35-p36 and provides further evidence of loci on chromosome 4q31 and 6q24.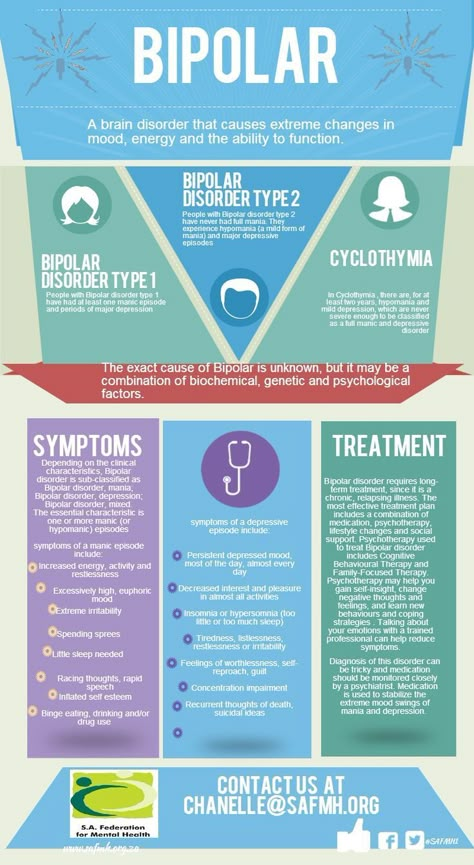 Am J Hum Genet. 2005;77:1102–1111. [PMC free article] [PubMed] [Google Scholar]
Am J Hum Genet. 2005;77:1102–1111. [PMC free article] [PubMed] [Google Scholar]
60. Zandi PP., Badner JA., Steele J., et al Genome-wide linkage scan of 98 bipolar pedigrees and analysis of clinical covariates. Mol Psychiatry. 2007;12:630–639. [PubMed] [Google Scholar]
61. Etain B., Mathieu F., Rietschel M., et al Genome-wide scan for genes involved in bipolar affective disorder in 70 European families ascertained through a bipolar type I early-onset proband: supportive evidence for linkage at 3p14. Mol Psychiatry. 2006;11:685–694. [PMC free article] [PubMed] [Google Scholar]
62. Badenhop RF., Moses MJ., Scimone A., et al Genetic refinement and physical mapping of a 2.3 Mb probable disease region associated with a bipolar affective disorder susceptibility locus on chromosome 4q35. Am J Med Genet B Neuropsychiatr Genet. 2003;1 17:23–32. [PubMed] [Google Scholar]
63. Dick DM., Foroud T., Edenberg HJ., et al Apparent replication of suggestive linkage on chromosome 16 in the NIMH genetics initiative bipolar pedigrees. Am J Med Genet. 2002;1 14:407–412. [PubMed] [Google Scholar]
Am J Med Genet. 2002;1 14:407–412. [PubMed] [Google Scholar]
64. Detera-Wadleigh SD., Badner JA., Yoshikawa T., et al Initial genome scan of the NIMH genetics initiative bipolar pedigrees: chromosomes 4:7:9:18:19:20, and 21q. Am J Med Genet. 1997;74:254–262. [PubMed] [Google Scholar]
65. Jones I., Hamshere M., Nangle JM., et al Bipolar affective puerperal psychosis: genome-wide significant evidence for linkage to chromosome 16. Am J Psychiatry. 2007;164:1099–1104. [PubMed] [Google Scholar]
66. Mclnnis MG., Lan TH., Willour VL., et al Genome-wide scan of bipolar disorder in 65 pedigrees: supportive evidence for linkage at 8q24:18q22:4q32:2p12, and 13q12. Mol Psychiatry. 2003;8:288–298. [PubMed] [Google Scholar]
67. Mclnnis MG., Dick DM., Willour VL., et al Genome-wide scan and conditional analysis in bipolar disorder: evidence for genomic interaction in the National Institute of Mental Health genetics initiative bipolar pedigrees. Biol Psychiatry. 2003;54:1265–1273. [PubMed] [Google Scholar]
Biol Psychiatry. 2003;54:1265–1273. [PubMed] [Google Scholar]
68. Foroud T., Castelluccio PF., Keller DL., et al Suggestive evidence of a locus on chromosome 10p using the NIMH genetics initiative bipolar affective disorder pedigrees. Am J Med Genet. 2000;96:18–23. [PubMed] [Google Scholar]
69. Ewald H., Flint T., Kruse TA., Mors O. A genome-wide scan shows significant linkage between bipolar disorder and chromosome 12q24.3 and suggestive linkage to chromosomes 1p22-21:4p16:6q14-22:10q26 and 16p13.3. Mol Psychiatry. 2002;15;7:734–744. [PubMed] [Google Scholar]
70. Cichon S., Schumacher J., Millier DJ., et al A genome screen for genes predisposing to bipolar affective disorder detects a new susceptibility locus on 8q. Hum Mol Genet. 2001;10:2933–2944. [PubMed] [Google Scholar]
71. Shink E., Morissette J., Sherrington R., Barden N. A genome-wide scan points to a susceptibility locus for bipolar disorder on chromosome 12. Moi Psychiatry. 2005;10:545–552. [PubMed] [Google Scholar]
2005;10:545–552. [PubMed] [Google Scholar]
72. Morissette J., Villeneuve A., Bordeleau L., et al Genome-wide search for linkage of bipolar affective disorders in a very large pedigree derived from a homogeneous population in quebec points to a locus of major effect on chromosome 12q23-q24. Am J Med Genet. 1999;88:567–587. [PubMed] [Google Scholar]
73. Kelsoe JR., Spence MA., Loetscher E., et al A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci USA. 2001;98:585–590. [PMC free article] [PubMed] [Google Scholar]
74. Badner JA., Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. [PubMed] [Google Scholar]
75. Potash JB., Zandi PP., Willour VL., et al Suggestive linkage to chromosomal regions 13q31 and 22q12 in families with psychotic bipolar disorder. Ami J Psychiatry. 2003;160:680–686. [PubMed] [Google Scholar]
76. Liu J., Juo SH., Terwilliger JD., et al A follow-up linkage study supports evidence for a bipolar affective disorder locus on chromosome 21q22. Am J Med Genet. 2001;105:189–194. [PubMed] [Google Scholar]
Liu J., Juo SH., Terwilliger JD., et al A follow-up linkage study supports evidence for a bipolar affective disorder locus on chromosome 21q22. Am J Med Genet. 2001;105:189–194. [PubMed] [Google Scholar]
77. Willour VL., Zandi PP., Huo Y., et al Genome scan of the fifty-six bipolar pedigrees from the NIMH genetics initiative replication sample: chromosomes 4:7:9:18:19:20, and 21. Am J Med Genet B Neuropsychiatr Genet. 2003;121:21–27. [PubMed] [Google Scholar]
78. Detera-Wadleigh SD., Badner JA., Goldin LR., et al Affected-sib-pair analyses reveal support of prior evidence for a susceptibility locus for bipolar disorder, on 21q. Ami J Hum Genet. 1996;58:1279–1285. [PMC free article] [PubMed] [Google Scholar]
79. Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999; 22:139–144. [PubMed] [Google Scholar]
80. Detera-Wadleigh SD., Liu CY., Maheshwari M., et al Sequence variation in DOCK9 and heterogeneity in bipolar disorder.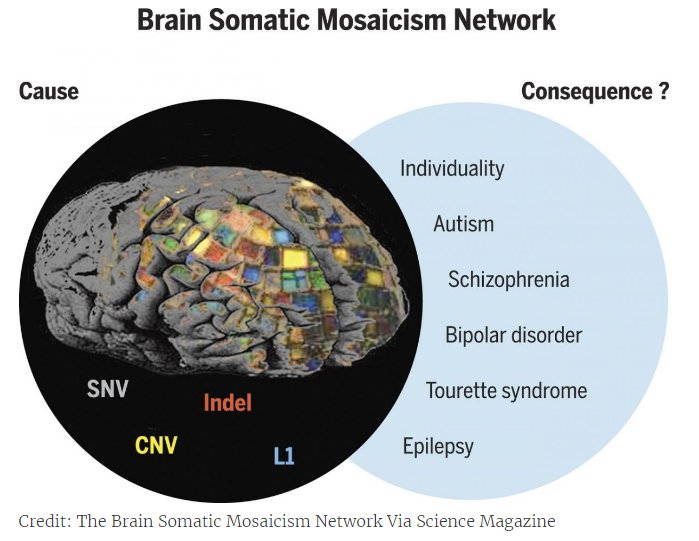 Psychiatr Genet. 2007;17:274–286. [PubMed] [Google Scholar]
Psychiatr Genet. 2007;17:274–286. [PubMed] [Google Scholar]
81. Mclnnes LA., Service SK., Reus VI., et al Fine-scale mapping of a locus for severe bipolar mood disorder on chromosome 18p11.3 in the Costa Rican population. Proc . Natl Acad Sci USA. 2001;98:11485–11490. [PMC free article] [PubMed] [Google Scholar]
82. Lee BD., Walss-Bass C., Thompson PM., et al Malic enzyme 2 and susceptibility to psychosis and mania. Psychiatry Res. 2007;150:1–11. [PubMed] [Google Scholar]
83. Ohnishi T., Yamada K., Ohba H., et al A promoter haplotype of the inositol monophosphatase 2 gene (IMPA2) at 18p11.2 confers a possible risk for bipolar disorder by enhancing transcription. Neuropsychopharmacology. 2007;32:1727–1737. [PubMed] [Google Scholar]
84. Washizuka S., Kametani M., Sasaki T., et al Association of mitochondrial complex I subunit gene NDUFV2 at 18p11 with schizophrenia in the Japanese population. Am J Med Genet B Neuropsychiatr Genet. 2006;141:301–304. [PubMed] [Google Scholar]
2006;141:301–304. [PubMed] [Google Scholar]
85. Weller AE., Dahl JP., Lohoff FW., Ferraro TN., Berrettini WH. Analysis of variations in the NAPG gene on chromosome 18p11 in bipolar disorder. Psychiatr Genet. 2006;16:3–8. [PubMed] [Google Scholar]
86. Barrett TB., Hauger RL., Kennedy JL., et al Evidence that a single nucleotide polymorphism in the promoter of the G protein receptor kinase 3 gene is associated with bipolar disorder. Mol Psychiatry: 2003;8:546–557. [PubMed] [Google Scholar]
87. Nievergelt CM., Kripke DF., Barrett TB., et al Suggestive evidence for association of the circadian genes PERIOD3 and ARNTL with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2006 May;141:234–241. [PMC free article] [PubMed] [Google Scholar]
88. Mansour HA., Wood J., Logue T., et al Association study of eight circadian genes with bipolar I disorder, schizoaffective disorder and schizophrenia. Genes. Brain Behav. 2006;5:150–157. [PubMed] [Google Scholar]
[PubMed] [Google Scholar]
89. Muglia P., Petronis A., Mundo E., Lander S., Cate T., Kennedy JL. Dopamine D4 receptor and tyrosine hydroxylase genes in bipolar disorder: evidence for a role of DRD4. Mol Psychiatry. 2002;7:860–866. [PubMed] [Google Scholar]
90. Del Zornpo M., De Luca V., Severino G., et al Haplotype association study between DRD1 gene and bipolar type I affective disorder in two samples from Canada and Sardinia. Am J Med Genet B Neuropsychiatr Genet. 2007;144:237–241. [PubMed] [Google Scholar]
91. Kirov G., Jones I., McCandless F., Craddock N., Owen MJ. Family-based association studies of bipolar disorder with candidate genes involved in dopamine neurotransmission: DBH, DAT1, COMT, DRD2, DRD3 and DRD5. Mol Psychiatry. 1999;4:558–565. [PubMed] [Google Scholar]
92. Neves FS., Silveira G., Romano-Silva MA., et al Is the 5-HTTLPR polymorphism associated with bipolar disorder or with suicidal behavior of bipolar disorder patients? Am J Med Genet B Neuropsychiatr Genet. 2008;147:114–116. [PubMed] [Google Scholar]
2008;147:114–116. [PubMed] [Google Scholar]
93. Ranade SS., Mansour H., Wood J., et al Linkage and association between serotonin 2A receptor gene polymorphisms and bipolar I disorder. Am J Med Genet B Neuropsychiatr Genet. 2003;121:28–34. [PubMed] [Google Scholar]
94. Kanazawa T., Glatt SJ., Kia-Keating B., Yoneda H., Tsuang MT. Meta-analysis reveals no association of the Val66Met polymorphism of brain-derived neurotrophic factor with either schizophrenia or bipolar disorder. Psychiatr Genet. 2007;17:165–170. [PubMed] [Google Scholar]
95. Atz ME., Rollins B., Vawter MP. NCAM1 association study of bipolar disorder and schizophrenia: polymorphisms and alternatively spliced isoforms lead to similarities and differences. Psychiatr Genet. 2007;17:55–67. [PMC free article] [PubMed] [Google Scholar]
96. Green EK., Raybould R., Macgregor S., et al Operation of the schizophrenia susceptibility gene, neuregulin 1, across traditional diagnostic boundaries to increase risk for bipolar disorder. Arch Gen Psychiatry. 2005;62:642–648. [PubMed] [Google Scholar]
Arch Gen Psychiatry. 2005;62:642–648. [PubMed] [Google Scholar]
97. Walss-Bass C., Raventos H., Montero AP., et al Association analyses of the neuregulin 1 gene with schizophrenia and manic psychosis in a Hispanic population. Acta Psychiatr Scand. 2006;113:314–321. [PubMed] [Google Scholar]
98. Detera-Wadleigh SD., McMahon FJ. G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol Psychiatry. 2006;60:106–114. [PubMed] [Google Scholar]
99. Chen YS., Akula N., Detera-Wadleigh SD., et al Findings in an independent sample support an association between bipolar affective disorder and the G72/G30 locus on chromosome 13q33. Mol Psychiatry. 2004;9:87–92. [PubMed] [Google Scholar]
100. GAIN Collaborative Research Group, Manolio TA., Rodriguez LL., et al New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39:1045–1051. [PubMed] [Google Scholar]
101. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. [PMC free article] [PubMed] [Google Scholar]
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. [PMC free article] [PubMed] [Google Scholar]
102. Shriner D., Vaughan LK., Padilla MA., Tiwari HK. Problems with genomewide association studies. Science. 2007;316:1840–1842. [PubMed] [Google Scholar]
103. Escamilla MA., Spesny M., Reus VI., et al Use of linkage disequilibrium approaches to map genes for bipolar disorder in the Costa Rican population. Am J Med Genet. 1996;67:244–253. [PubMed] [Google Scholar]
104. Escamilla MA., Mclnnes LA., Spesny M., et al Assessing the feasibility of linkage disequilibrium methods for mapping complex traits: an initial screen for bipolar disorder loci on chromosome. K.Am J Hum Genet. 1999;64:1670–1678. [PMC free article] [PubMed] [Google Scholar]
105. phoff RA., Escamilla MA., Service SK., et al Genome-wide linkage disequilibrium mapping of severe bipolar disorder in a population isolate. Am J Hum Genet. 2002;71:565–574. [PMC free article] [PubMed] [Google Scholar]
Am J Hum Genet. 2002;71:565–574. [PMC free article] [PubMed] [Google Scholar]
106. Baum AE., Akula N., Cabanero M., et al A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. [PMC free article] [PubMed] [Google Scholar]
107. Gottesman II., Shields J. Genetic theorizing and schizophrenia. Br J Psychiatry. 1973;122:15–30. [PubMed] [Google Scholar]
108. Ôzer S., Ayhan Y., Uluflahin A. The utility of an endophenotype approach in overcoming the difficulties in bipolar and schizophrenia genetics. Turk J Psychiatry. 2004;15:125–137. [PubMed] [Google Scholar]
109. First MB., Pincus HA., Levine JB., Williams JB., Ustun B., Peele R. Clinical utility as a criterion for revising psychiatric diagnoses. Am J Psychiatry. 2004;161:946–954. [PubMed] [Google Scholar]
110. Gershon ES., Goldin LR. Clinical methods in psychiatric genetics, I: robustness of genetic marker investigative strategies. Acta Psychiatr Scand. 1986;74:113–118. [PubMed] [Google Scholar]
Acta Psychiatr Scand. 1986;74:113–118. [PubMed] [Google Scholar]
111. Leboyer M., Belliver F., Nosten-Bertrand M., Jouvent R., Pauls D., Mallet J. Psychiatric genetics: search for phenotypes. Trends Neurosci. 1998;21:102–105. [PubMed] [Google Scholar]
112. MacDonald C., Bullmore ET., Sham PC., Chintus X., Wickam H., Bramon E., et al Association of genetic risks for schizophrenia and bipolar disorder with specific and generic brain structural endophenotypes. Arch Gen Psychiatry. 2004;61:974–984. [PubMed] [Google Scholar]
113. Altshuller LL., Curran JG., Hauser P., Mintz J., Denicoff K., Post R. T2hyperintensities in bipolar disorder: magnetic resonance imaging comparison and literature meta-analysis. Am J Psychiatry. 1995;152:1139–1144. [PubMed] [Google Scholar]
114. Videbech P. MRI findings in patients with affective disorder: A metaanalysis. Acta Psychiatr Scand. 1997;96:157–168. [PubMed] [Google Scholar]
115. MacDonald C. , Zenelli J., Rabe-Hesketh S., et al Meta-analysis of magnetic resonance imaging brain morphometry studies in bipolar disorder. Biol Psychiatry. 2004;56:411–417. [PubMed] [Google Scholar]
, Zenelli J., Rabe-Hesketh S., et al Meta-analysis of magnetic resonance imaging brain morphometry studies in bipolar disorder. Biol Psychiatry. 2004;56:411–417. [PubMed] [Google Scholar]
116. Clark L., Iverson SD., Goodwin GM. Sustained attention deficit in bipolar disorder. Br J Psychiatry. 2002;180:313–319. [PubMed] [Google Scholar]
117. Seidman LJ., Kremen WS., Koren D., Faraone SV., Goldstein JM., Tsuang MT. A comparative profile analysis of neuropsychological functioning in patients with schizophrenia and bipolar psychosis. Schizophr Res. 2002;53:31–44. [PubMed] [Google Scholar]
118. Sobczak S., Honig A., Nicolson NA., Riedel WJ. Effects of acute tryptophan depletion on mood and Cortisol release in first-degree relatives of type I and type II bipolar patients and healthy matched controls. Neuropsychopharmacoiogy. 2002;27: 834–842. [PubMed] [Google Scholar]
119. Gould TD., Chen G., Manji HK. In vivo evidence in the brain for lithium inhibition of glycogen synthase kinase-3. Neuropsychopharmacoiogy. 2004;29:32–38. [PubMed] [Google Scholar]
Neuropsychopharmacoiogy. 2004;29:32–38. [PubMed] [Google Scholar]
120. Klein PS., Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci U S A. 1996;93:8455–8459. [PMC free article] [PubMed] [Google Scholar]
121. Benadetti F., Bernasconi A., Lorenzi C., et al A single nucleotide polymorphism in glycogen synthase kinase-3 beta promoter gene influences onset of illness in patients affected by bipolar disorder. Neurosci Lett. 2004;355:37–40. [PubMed] [Google Scholar]
122. Glahn DC., Bearden CE., Niendam TA., Escamilla MA. The feasibility of neuropsychological endophenotypes in the search for genes associated with bipolar affective disorder. Bipolar Disord. 2004;6:171–182. [PubMed] [Google Scholar]
123. Savitz JB., Ramesar RS. Personality: is it a viable endophenotype for genetic studies of bipolar affective disorder? Bipolar Disord. 2006;8:322–337. [PubMed] [Google Scholar]
124. Escamilla MA. Population isolates: their special value for locating genes for bipolar disorder. Bipolar Disord. 2001;3:299–317. [PubMed] [Google Scholar]
Escamilla MA. Population isolates: their special value for locating genes for bipolar disorder. Bipolar Disord. 2001;3:299–317. [PubMed] [Google Scholar]
125. Sullivan PF., Neale BM., van den Oord E., Miles MF., Neale MC., Bulik CM., Joyce PR., Straub RE., Kendler KS. Candidate genes for nicotine dependence via linkage, epistasis, and bioinformatics. Am J Med Genet B Neuropsychiatr Genet. 2004;126:23–36. [PubMed] [Google Scholar]
126. Kelsoe JR., Niculescu AB 3rd. Finding genes for bipolar disorder in the functional genomics era: from convergent functional genomics to phenomics and back. CNS Spectr. 2002;7:215–216. 223–226. [PubMed] [Google Scholar]
127. Conn RD., Van Erp C., Habashi JP., et al Angiotensin II type 1 receptor blockade attenuates TGF-beta-induced failure of muscle regeneration in multiple myopathic states. Nat Med. 2007;13:204–210. [PMC free article] [PubMed] [Google Scholar]
128. Stevens A., Price J. Evolutionary Psychiatry. London, UK: Routledge:1996. [Google Scholar]
London, UK: Routledge:1996. [Google Scholar]
Genetics of bipolar disorder - PMC
1. National Institute of Mental Health Bipolar disorder among adults. [Accessed January 10, 2014]. Available from: http://www.nimh.nih.gov/statistics/1bipolar_adult.shtml.
2. American Psychiatric Association [homepage on the internet] [Accessed January 10, 2014]. Available from: http://www.psychiatry.org/home.
3. Merikangas KR, Cui L, Heaton L, et al. Independence of familial transmission of mania and depression: results of the NIMH family study of affective spectrum disorders. Mol Psychiatry. 2013 Oct 15; Epub. [PubMed] [Google Scholar]
4. Vandeleur CL, Merikangas KR, Strippoli MP, Castelao E, Preisig M. Specificity of psychosis, mania and major depression in a contemporary family study. Mol Psychiatry. 2013 Oct 15; Epub. [PubMed] [Google Scholar]
5. Corry J, Green M, Roberts G, et al. Anxiety, stress and perfectionism in bipolar disorder. J Affect Disord. 2013;151(3):1016–1024. [PubMed] [Google Scholar]
2013;151(3):1016–1024. [PubMed] [Google Scholar]
6. van Emmerik-van Oortmerssen K, van de Glind G, Koeter MW, et al. Psychiatric comorbidity in treatment seeking substance use disorder patients with and without ADHD; results of the IASP study. Addiction. 2013 Oct 4; Epub. [PMC free article] [PubMed] [Google Scholar]
7. Di Florio A, Craddock N, van den Bree M. Alcohol misuse in bipolar disorder. A systematic review and meta-analysis of comorbidity rates. Eur Psychiatry. 2013 Sep 25; Epub. [PubMed] [Google Scholar]
8. Di Nicola M, Sala L, Romo L, et al. Adult attention-deficit/hyperactivity disorder in major depressed and bipolar subjects: role of personality traits and clinical implications. Eur Arch Psychiatry Clin Neurosci. 2013 Sep 28; Epub. [PubMed] [Google Scholar]
9. Leboyer M, Kupfer DJ. Bipolar disorder: new perspectives in health care and prevention. J Clin Psychiatry. 2010;71(12):1689–1695. [PMC free article] [PubMed] [Google Scholar]
10. Fagiolini A, Kupfer DJ, Rucci P, Scott JA, Novick DM, Frank E. Suicide attempts and ideation in patients with bipolar I disorder. J Clin Psychiatry. 2004;65(4):509–514. [PubMed] [Google Scholar]
Suicide attempts and ideation in patients with bipolar I disorder. J Clin Psychiatry. 2004;65(4):509–514. [PubMed] [Google Scholar]
11. Hauser M, Galling B, Correll CU. Suicidal ideation and suicide attempts in children and adolescents with bipolar disorder: a systematic review of prevalence and incidence rates, correlates, and targeted interventions. Bipolar Disord. 2013;15(5):507–523. [PMC free article] [PubMed] [Google Scholar]
12. Post RM, Leverich GS, Kupka RW, et al. Early-onset bipolar disorder and treatment delay are risk factors for poor outcome in adulthood. J Clin Psychiatry. 2010;71(7):864–872. [PubMed] [Google Scholar]
13. Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123C(1):48–58. [PubMed] [Google Scholar]
14. Wray NR, Gottesman II. Using summary data from the Danish national registers to estimate heritabilities for schizophrenia, bipolar disorder, and major depressive disorder. Front Genet. 2012;3:118. [PMC free article] [PubMed] [Google Scholar]
Front Genet. 2012;3:118. [PMC free article] [PubMed] [Google Scholar]
15. Aas M, Etain B, Bellivier F, et al. Additive effects of childhood abuse and cannabis abuse on clinical expressions of bipolar disorders. Psychol Med. 2013 Sep 13; Epub. [PubMed] [Google Scholar]
16. Brown AS. The environment and susceptibility to schizophrenia. Prog Neurobiol. 2011;93(1):23–58. [PMC free article] [PubMed] [Google Scholar]
17. Meli G, Ottl B, Paladini A, Cataldi L. Prenatal and perinatal risk factors of schizophrenia. J Matern Fetal Neonatal Med. 2012;25(12):2559–2563. [PubMed] [Google Scholar]
18. Canetta SE, Brown AS. Prenatal infection, maternal immune activation, and risk for schizophrenia. Transl Neurosci. 2012;3(4):320–327. [PMC free article] [PubMed] [Google Scholar]
19. Khandaker GM, Zimbron J, Lewis G, Jones PB. Prenatal maternal infection, neurodevelopment and adult schizophrenia: a systematic review of population-based studies. Psychol Med. 2013;43(2):239–257. [PMC free article] [PubMed] [Google Scholar]
[PMC free article] [PubMed] [Google Scholar]
20. Haukvik UK, McNeil T, Lange EH, et al. Pre- and perinatal hypoxia associated with hippocampus/amygdala volume in bipolar disorder. Psychol Med. 2013;27:1–11. [PMC free article] [PubMed] [Google Scholar]
21. Kenny EM, Cormican P, Furlong S, et al. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol Psychiatry. 2013 Oct 15; Epub. [PubMed] [Google Scholar]
22. Craddock N, Sklar P. Genetics of bipolar disorder. Lancet. 2013;381(9878):1654–1662. [PubMed] [Google Scholar]
23. Szczepankiewicz A. Evidence for single nucleotide polymorphisms and their association with bipolar disorder. Neuropsychiatr Dis Treat. 2013;9:1573–1582. [PMC free article] [PubMed] [Google Scholar]
24. Sklar P, Smoller JW, Fan J, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13(6):558–569. [PMC free article] [PubMed] [Google Scholar]
25. Ferreira MA, O‘Donovan MC, Meng YA, et al. Wellcome Trust Case Control Consortium. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40(9):1056–1058. [PMC free article] [PubMed] [Google Scholar]
Wellcome Trust Case Control Consortium. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40(9):1056–1058. [PMC free article] [PubMed] [Google Scholar]
26. Moskvina V, Craddock N, Holmans P, et al. Gene-wide analyses of genome-wide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol Psychiatry. 2009;14(3):252–260. [PMC free article] [PubMed] [Google Scholar]
27. Kloiber S, Czamara D, Karbalai N, et al. ANK3 and CACNA1C: missing genetic link for bipolar disorder and major depressive disorder in two German case-control samples. J Psychiatr Res. 2012;46(8):973–979. [PubMed] [Google Scholar]
28. Zhang J, Cai J, Zhang X, et al. Does the bipolar disorder-associated CACNA1C gene confer susceptibility to schizophrenia in Han Chinese? J Mol Neurosci. 2013;51(2):474–477. [PubMed] [Google Scholar]
29. Zhang X, Zhang C, Wu Z, et al. Association of genetic variation in CACNA1C with bipolar disorder in Han Chinese. J Affect Disord. 2013;150(2):261–265. [PubMed] [Google Scholar]
Association of genetic variation in CACNA1C with bipolar disorder in Han Chinese. J Affect Disord. 2013;150(2):261–265. [PubMed] [Google Scholar]
30. Gershon ES, Grennan K, Busnello J, et al. A rare mutation of CACNA1C in a patient with bipolar disorder, and decreased gene expression associated with a bipolar-associated common SNP of CACNA1C in brain. Mol Psychiatry. 2013 Aug 27; Epub. [PMC free article] [PubMed] [Google Scholar]
31. Bigos KL, Mattay VS, Callicott JH, et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch Gen Psychiatry. 2010;67(9):939–945. [PMC free article] [PubMed] [Google Scholar]
32. Wang F, McIntosh AM, He Y, Gelernter J, Blumberg HP. The association of genetic variation in CACNA1C with structure and function of a frontotemporal system. Bipolar Disord. 2011;13(7–8):696–700. [PMC free article] [PubMed] [Google Scholar]
33. Soeiro-de-Souza MG, Otaduy MC, Dias CZ, Bio DS, Machado-Vieira R, Moreno RA. The impact of the CACNA1C risk allele on limbic structures and facial emotions recognition in bipolar disorder subjects and healthy controls. J Affect Disord. 2012;141(1):94–101. [PubMed] [Google Scholar]
J Affect Disord. 2012;141(1):94–101. [PubMed] [Google Scholar]
34. Soeiro-de-Souza MG, Bio DS, Dias VV, Vieta E, Machado-Vieira R, Moreno RA. The CACNA1C risk allele selectively impacts on executive function in bipolar type I disorder. Acta Psychiatr Scand. 2013;128(5):362–369. [PubMed] [Google Scholar]
35. Zhang Q, Shen Q, Xu Z, et al. The effects of CACNA1C gene polymorphism on spatial working memory in both healthy controls and patients with schizophrenia or bipolar disorder. Neuropsychopharmacology. 2012;37(3):677–684. [PMC free article] [PubMed] [Google Scholar]
36. Paulus FM, Bedenbender J, Krach S, et al. Association of rs1006737 in CACNA1C with alterations in prefrontal activation and fronto-hippocampal connectivity. Hum Brain Mapp. 2013 Feb 13; Epub. [PMC free article] [PubMed] [Google Scholar]
37. Cross-Disorder Group of the Psychiatric Genomics Consortium. Smoller JW, Craddock N, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–1379. [PMC free article] [PubMed] [Google Scholar]
Lancet. 2013;381(9875):1371–1379. [PMC free article] [PubMed] [Google Scholar]
38. Green EK, Hamshere M, Forty L, et al. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case-control sample. Mol Psychiatry. 2013;18(12):1302–1307. [PMC free article] [PubMed] [Google Scholar]
39. Hamshere ML, Walters JT, Smith R, et al. Genome-wide significant associations in schizophrenia to ITIh4/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. 2013;18(6):708–712. [PMC free article] [PubMed] [Google Scholar]
40. Splawski I, Timothy KW, Sharpe LM, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119(1):19–31. [PubMed] [Google Scholar]
41. Hsiao PY, Tien HC, Lo CP, Juang JM, Wang YH, Sung RJ. Gene mutations in cardiac arrhythmias: a review of recent evidence in ion channelopathies. Appl Clin Genet. 2013;6:1–13. [PMC free article] [PubMed] [Google Scholar]
Appl Clin Genet. 2013;6:1–13. [PMC free article] [PubMed] [Google Scholar]
42. Boczek NJ, Best JM, Tester DJ, et al. Exome sequencing and systems biology converge to identify novel mutations in the L-type calcium channel, CACNA1C, linked to autosomal dominant long QT syndrome. Circ Cardiovasc Genet. 2013;6(3):279–289. [PMC free article] [PubMed] [Google Scholar]
43. Brugada J, Brugada P. Further characterization of the syndrome of right bundle branch block, ST segment elevation, and sudden cardiac death. J Cardiovasc Electrophysiol. 1997;8(3):325–331. [PubMed] [Google Scholar]
44. Dao DT, Mahon PB, Cai X, et al. Mood disorder susceptibility gene CACNA1C modifies mood-related behaviors in mice and interacts with sex to influence behavior in mice and diagnosis in humans. Biol Psychiatry. 2010;68(9):801–810. [PMC free article] [PubMed] [Google Scholar]
45. PubMed [homepage on the internet] [Accessed January 10, 2014]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/
46. Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43(10):977–983. [PMC free article] [PubMed] [Google Scholar]
Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43(10):977–983. [PMC free article] [PubMed] [Google Scholar]
47. Tesli M, Egeland R, Sønderby IE, et al. No evidence for association between bipolar disorder risk gene variants and brain structural phenotypes. J Affect Disord. 2013;151(1):291–297. [PubMed] [Google Scholar]
48. Heinrich A, Lourdusamy A, Tzschoppe J, et al. The risk variant in ODZ4 for bipolar disorder impacts on amygdala activation during reward processing. Bipolar Disord. 2013 Apr 24; Epub. [PubMed] [Google Scholar]
49. Tesli M, Skatun KC, Ousdal OT, et al. CACNA1C risk variant and amygdala activity in bipolar disorder, schizophrenia and healthy controls. PLoS One. 2013;8(2):e56970. [PMC free article] [PubMed] [Google Scholar]
50. Krug A, Witt SH, Backes H, et al. A genome-wide supported variant in CACNA1C influences hippocampal activation during episodic memory encoding and retrieval. Eur Arch Psychiatry Clin Neurosci. 2013 Jul 17; Epub. [PubMed] [Google Scholar]
Eur Arch Psychiatry Clin Neurosci. 2013 Jul 17; Epub. [PubMed] [Google Scholar]
51. Ben-Zur T, Feige E, Motro B, Wides R. The mammalian Odz gene family: homologs of a Drosophila pair-rule gene with expression implying distinct yet overlapping developmental roles. Dev Biol. 2000;217(1):107–120. [PubMed] [Google Scholar]
52. Zhou XH, Brandau O, Feng K, et al. The murine Ten-m/Odz genes show distinct but overlapping expression patterns during development and in adult brain. Gene Expr Patterns. 2003;3(4):397–405. [PubMed] [Google Scholar]
53. Suzuki N, Fukushi M, Kosaki K, et al. Teneurin-4 is a novel regulator of oligodendrocyte differentiation and myelination of small-diameter axons in the CNS. J Neurosci. 2012;32(34):11586–11599. [PMC free article] [PubMed] [Google Scholar]
54. Cichon S, Mühleisen TW, Degenhardt FA, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88(3):372–381.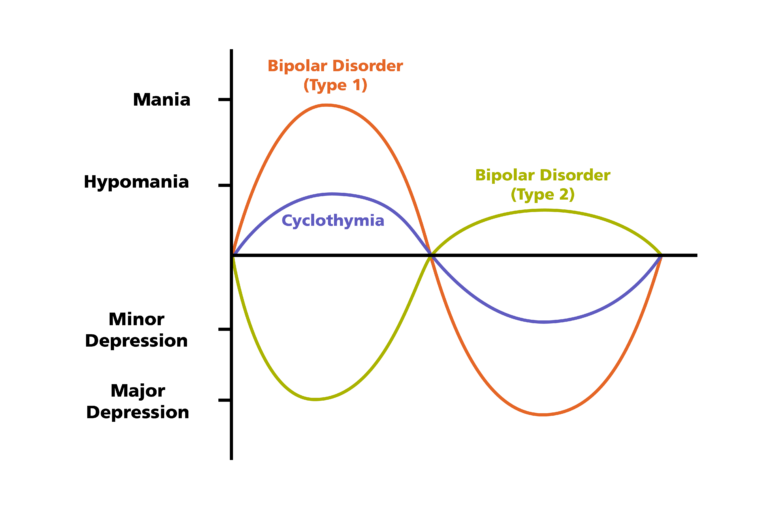 [PMC free article] [PubMed] [Google Scholar]
[PMC free article] [PubMed] [Google Scholar]
55. Oruč L, Kapur-Pojskić L, Ramić J, Pojskić N, Bajrović K. Assessment of relatedness between neurocan gene as bipolar disorder susceptibility locus and schizophrenia. Bosn J Basic Med Sci. 2012;12(4):245–248. [PMC free article] [PubMed] [Google Scholar]
56. Mühleisen TW, Mattheisen M, Strohmaier J, et al. Association between schizophrenia and common variation in neurocan (NCAN), a genetic risk factor for bipolar disorder. Schizophr Res. 2012;138(1):69–73. [PubMed] [Google Scholar]
57. Schultz CC, Mühleisen TW, Nenadic I, et al. Common variation in NCAN, a risk factor for bipolar disorder and schizophrenia, influences local cortical folding in schizophrenia. Psychol Med. 2013 Jun 24; Epub. [PubMed] [Google Scholar]
58. Zhou XH, Brakebusch C, Matthies H, et al. Neurocan is dispensable for brain development. Mol Cell Biol. 2001;21(17):5970–5978. [PMC free article] [PubMed] [Google Scholar]
59. Rauch U, Zhou XH, Roos G. Extracellular matrix alterations in brains lacking four of its components. Biochem Biophys Res Commun. 2005;328(2):608–617. [PubMed] [Google Scholar]
Extracellular matrix alterations in brains lacking four of its components. Biochem Biophys Res Commun. 2005;328(2):608–617. [PubMed] [Google Scholar]
60. Lu Y, Feskens EJ, Boer JM, et al. Exploring genetic determinants of plasma total cholesterol levels and their predictive value in a longitudinal study. Atherosclerosis. 2010;213(1):200–205. [PubMed] [Google Scholar]
61. Yan TT, Yin RX, Li Q, et al. Sex-specific association of rs16996148 SNP in the NCAN/CILP2/PBX4 and serum lipid levels in the Mulao and Han populations. Lipids Health Dis. 2011;10:248. [PMC free article] [PubMed] [Google Scholar]
62. Zhou L, Ding H, Zhang X, et al. Genetic variants at newly identified lipid loci are associated with coronary heart disease in a Chinese Han population. PLoS One. 2011;6(11):e27481. [PMC free article] [PubMed] [Google Scholar]
63. Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7(3):e1001324. [PMC free article] [PubMed] [Google Scholar]
PLoS Genet. 2011;7(3):e1001324. [PMC free article] [PubMed] [Google Scholar]
64. Gorden A, Yang R, Yerges-Armstrong LM, et al. Genetic variation at NCAN locus is associated with inflammation and fibrosis in nonalcoholic fatty liver disease in morbid obesity. Hum Hered. 2013;75(1):34–43. [PMC free article] [PubMed] [Google Scholar]
65. Nakayama K, Bayasgalan T, Yamanaka K, et al. Large scale replication analysis of loci associated with lipid concentrations in a Japanese population. J Med Genet. 2009;46(6):370–374. [PubMed] [Google Scholar]
66. [Accessed January 10, 2014];Online Mendelian Inheritance in Man (OMIM) [homepage on the internet] Available from: http://www.ncbi.nlm.nih.gov/omim. [Google Scholar]
67. Boer H, Holland A, Whittington J, Butler J, Webb T, Clarke D. Psychotic illness in people with Prader Willi syndrome due to chromosome 15 maternal uniparental disomy. Lancet. 2002;359(9301):135–136. [PubMed] [Google Scholar]
68. Burnside RD, Pasion R, Mikhail FM, et al.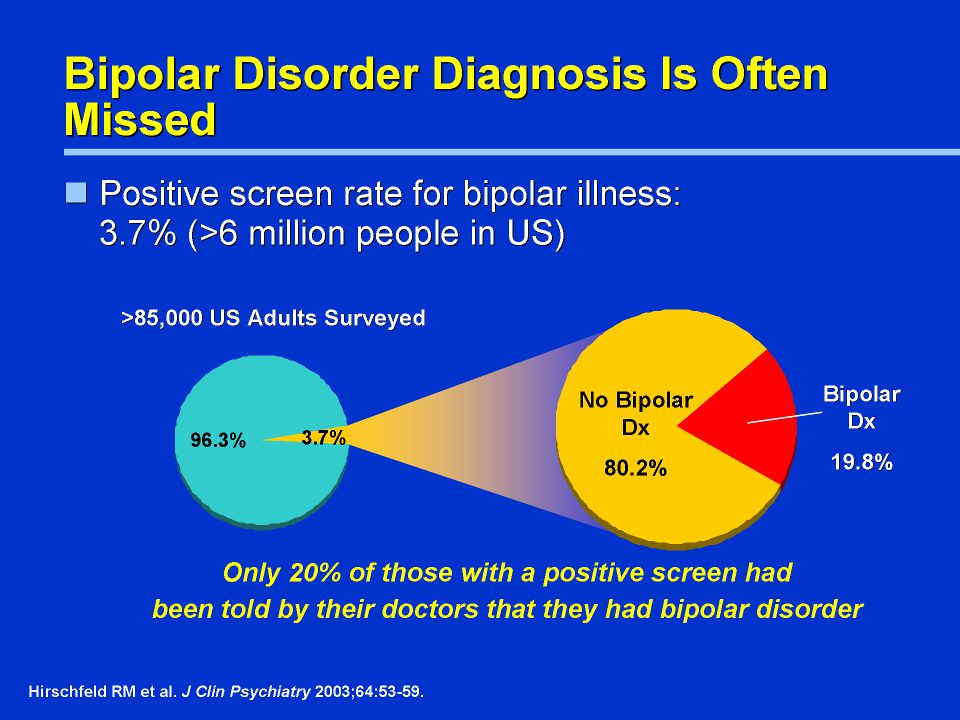 Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: a susceptibility region for neurological dysfunction including developmental and language delay. Hum Genet. 2011;130(4):517–528. [PMC free article] [PubMed] [Google Scholar]
Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: a susceptibility region for neurological dysfunction including developmental and language delay. Hum Genet. 2011;130(4):517–528. [PMC free article] [PubMed] [Google Scholar]
69. Skokauskas N, Sweeny E, Meehan J, Gallagher L. Mental health problems in children with Prader-Willi syndrome. J Can Acad Child Adolesc Psychiatry. 2012;21(3):194–203. [PMC free article] [PubMed] [Google Scholar]
70. Fan Z, Greenwood R, Fisher A, Pendyal S, Powell CM. Characteristics and frequency of seizure disorder in 56 patients with Prader-Willi syndrome. Am J Med Genet A. 2009;149A(7):1581–1584. [PubMed] [Google Scholar]
71. Sutcliffe JS, Nakao M, Christian S, et al. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8(1):52–58. [PubMed] [Google Scholar]
72. Hall BD, Smith DW. Prader-Willi syndrome. A resumé of 32 cases including an instance of affected first cousins, one of whom is of normal stature and intelligence. J Pediatr. 1972;81(2):286–293. [PubMed] [Google Scholar]
J Pediatr. 1972;81(2):286–293. [PubMed] [Google Scholar]
73. Autio S, Pihko H, Tengström C. Clinical features in a de novo interstitial deletion 15q13 to q15. Clin Genet. 1988;34(5):293–298. [PubMed] [Google Scholar]
74. Ninomiya S, Yokoyama Y, Kawakami M, Une T, Maruyama H, Morishima T. Unique maternal deletion of 15q in a patient with some symptoms of Prader-Willi syndrome. Pediatr Int. 2005;47(5):541–545. [PubMed] [Google Scholar]
75. Sharp AJ, Mefford HC, Li K, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40(3):322–328. [PMC free article] [PubMed] [Google Scholar]
76. Helbig I, Mefford HC, Sharp AJ, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41(2):160–162. [PMC free article] [PubMed] [Google Scholar]
77. de Kovel CG, Trucks H, Helbig I, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133(Pt 1):23–32. [PMC free article] [PubMed] [Google Scholar]
Brain. 2010;133(Pt 1):23–32. [PMC free article] [PubMed] [Google Scholar]
78. van Bon BW, Mefford HC, Menten B, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009;46(8):511–523. [PMC free article] [PubMed] [Google Scholar]
79. Miller DT, Shen Y, Weiss LA, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46(4):242–248. [PMC free article] [PubMed] [Google Scholar]
80. Pagnamenta AT, Wing K, Sadighi Akha E, et al. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet. 2009;17(5):687–692. [PMC free article] [PubMed] [Google Scholar]
81. Ben-Shachar S, Lanpher B, German JR, et al. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet. 2009;46(6):382–388. [PMC free article] [PubMed] [Google Scholar]
82. Sahoo T, Theisen A, Rosenfeld JA, et al. Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med. 2011;13(10):868–880. [PubMed] [Google Scholar]
Sahoo T, Theisen A, Rosenfeld JA, et al. Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med. 2011;13(10):868–880. [PubMed] [Google Scholar]
83. Shinawi M, Schaaf CP, Bhatt SS, et al. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat Genet. 2009;41(12):1269–1271. [PMC free article] [PubMed] [Google Scholar]
84. Hoppman-Chaney N, Wain K, Seger PR, Superneau DW, Hodge JC. Identification of single gene deletions at 15q13.3: further evidence that CHRNA7 causes the 15q13.3 microdeletion syndrome phenotype. Clin Genet. 2013;83(4):345–351. [PubMed] [Google Scholar]
85. Endris V, Hackmann K, Neuhann TM, et al. Homozygous loss of CHRNA7 on chromosome 15q13.3 causes severe encephalopathy with seizures and hypotonia. Am J Med Genet A. 2010;152A(11):2908–2911. [PubMed] [Google Scholar]
86. Pagon Roberta A, Adam Margaret P, Bird Thomas D, Dolan Cynthia R, Fong Chin-To, Smith Richard JH, Stephens and Karen. , editors. GeneReviews™ [internet] Seattle (WA): University of Washington, Seattle; 1993–2014. [Accessed October 18, 2013]. Available from http://www.ncbi.nlm.nih.gov/books/ [Google Scholar]
, editors. GeneReviews™ [internet] Seattle (WA): University of Washington, Seattle; 1993–2014. [Accessed October 18, 2013]. Available from http://www.ncbi.nlm.nih.gov/books/ [Google Scholar]
87. Mehta L, Duckett DP, Young ID. Behaviour disorder in monosomy 10qter. J Med Genet. 1987;24(3):185–186. [PMC free article] [PubMed] [Google Scholar]
88. Courtens W, Wuyts W, Rooms L, Pera SB, Wauters J. A subterminal deletion of the long arm of chromosome 10: a clinical report and review. Am J Med Genet A. 2006;140(4):402–409. [PubMed] [Google Scholar]
89. Miller ND, Nance MA, Wohler ES, et al. Molecular (SNP) analyses of overlapping hemizygous deletions of 10q25.3 to 10qter in four patients: evidence for HMX2 and HMX3 as candidate genes in hearing and vestibular function. Am J Med Genet A. 2009;149A(4):669–680. [PMC free article] [PubMed] [Google Scholar]
90. Yatsenko SA, Kruer MC, Bader PI, et al. Identification of critical regions for clinical features of distal 10q deletion syndrome. Clin Genet. 2009;76(1):54–62. [PubMed] [Google Scholar]
Clin Genet. 2009;76(1):54–62. [PubMed] [Google Scholar]
91. Irving M, Hanson H, Turnpenny P, et al. Deletion of the distal long arm of chromosome 10; is there a characteristic phenotype? A report of 15 de novo and familial cases. Am J Med Genet A. 2003;123A(2):153–163. [PubMed] [Google Scholar]
92. Bassett AS, Chow EW, Husted J, et al. Clinical features of 78 adults with 22q11 deletion syndrome. Am J Med Genet A. 2005;138(4):307–313. [PMC free article] [PubMed] [Google Scholar]
93. McLean SD, Saal HM, Spinner NB, Emanuel BS, Driscoll DA. Velo-cardio-facial syndrome. Intrafamilial variability of the phenotype. Am J Dis Child. 1993;147(11):1212–1216. [PubMed] [Google Scholar]
94. Jolin EM, Weller RA, Weller EB. Occurrence of affective disorders compared to other psychiatric disorders in children and adolescents with 22q11.2 deletion syndrome. J Affect Disord. 2012;136(3):222–228. [PubMed] [Google Scholar]
95. Aneja A, Fremont WP, Antshel KM, et al. Manic symptoms and behavioral dysregulation in youth with velocardiofacial syndrome (22q11. 2 deletion syndrome) J Child Adolesc Psychopharmacol. 2007;17(1):105–114. [PubMed] [Google Scholar]
2 deletion syndrome) J Child Adolesc Psychopharmacol. 2007;17(1):105–114. [PubMed] [Google Scholar]
96. Shprintzen RJ, Goldberg RB, Young D, Wolford L. The velo-cardio-facial syndrome: a clinical and genetic analysis. Pediatrics. 1981;67(2):167–172. [PubMed] [Google Scholar]
97. Shprintzen RJ. Velo-cardio-facial syndrome: 30 Years of study. Dev Disabil Res Rev. 2008;14(1):3–10. [PMC free article] [PubMed] [Google Scholar]
98. Evers LJ, Vermaak MP, Engelen JJ, Curfs LM. The velocardiofacial syndrome in older age: dementia and autistic features. Genet Couns. 2006;17(3):333–340. [PubMed] [Google Scholar]
99. Butcher NJ, Chow EW, Costain G, Karas D, Ho A, Bassett AS. Functional outcomes of adults with 22q11.2 deletion syndrome. Genet Med. 2012;14(10):836–843. [PMC free article] [PubMed] [Google Scholar]
100. De Leersnyder H. Smith-Magenis syndrome. Handb Clin Neurol. 2013;111:295–2966. [PubMed] [Google Scholar]
101. Girirajan S, Elsas LJ, 2nd, Devriendt K, Elsea SH. RAI1 variations in Smith-Magenis syndrome patients without 17p11.2 deletions. J Med Genet. 2005;42(11):820–828. [PMC free article] [PubMed] [Google Scholar]
RAI1 variations in Smith-Magenis syndrome patients without 17p11.2 deletions. J Med Genet. 2005;42(11):820–828. [PMC free article] [PubMed] [Google Scholar]
102. Elsea SH, Williams SR. Smith-Magenis syndrome: haploinsufficiency of RAI1 results in altered gene regulation in neurological and metabolic pathways. Expert Rev Mol Med. 2011;13:e14. [PubMed] [Google Scholar]
103. Juyal RC, Figuera LE, Hauge X, et al. Molecular analyses of 17p11.2 deletion in 62 Smith-Magenis syndrome patients. Am J Med Genet. 1996;58:998–1007. [PMC free article] [PubMed] [Google Scholar]
104. Gershon ES, Alliey-Rodriguez N. New ethical issues for genetic counseling in common mental disorders. Am J Psychiatry. 2013;170(9):968–976. [PubMed] [Google Scholar]
105. Wilfond BS, Thomson EJ. Models of public health genetic policy development. In: Khoury MJ, Burke W, Thomson EJ, editors. Genetics and Public Health in the 21th Century. Oxford, UK: Oxford University Press; 2000. pp. 61–81. [Google Scholar]
Genetics of Bipolar Disorders - PsyAndNeuro.
 ru
ru
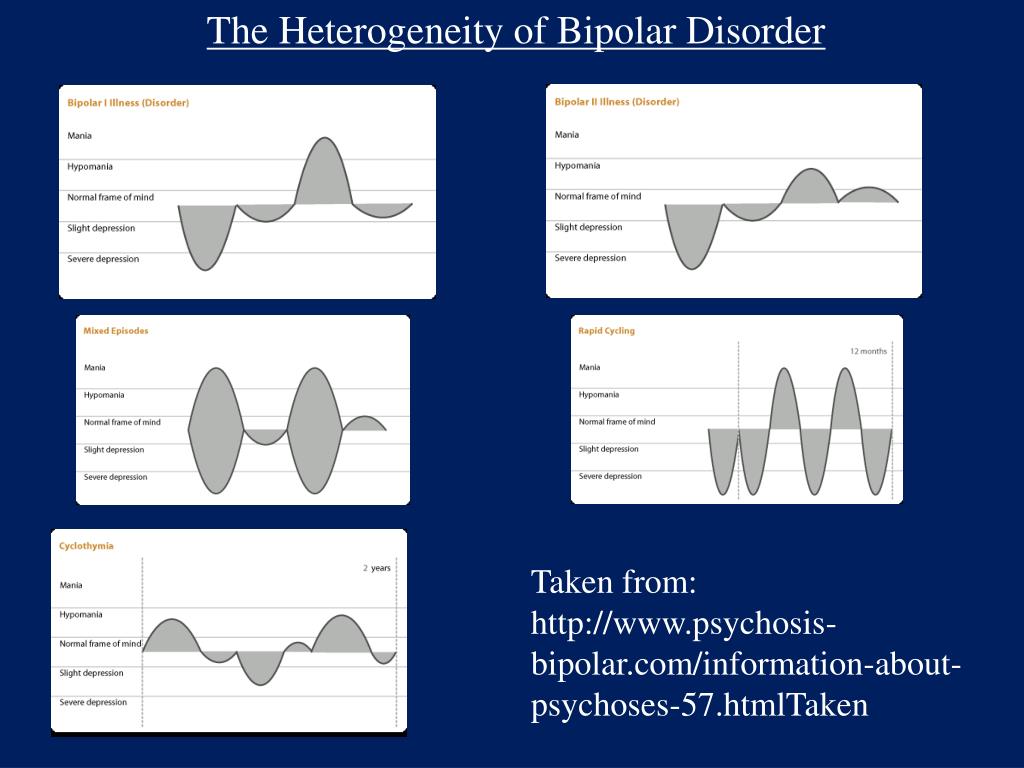
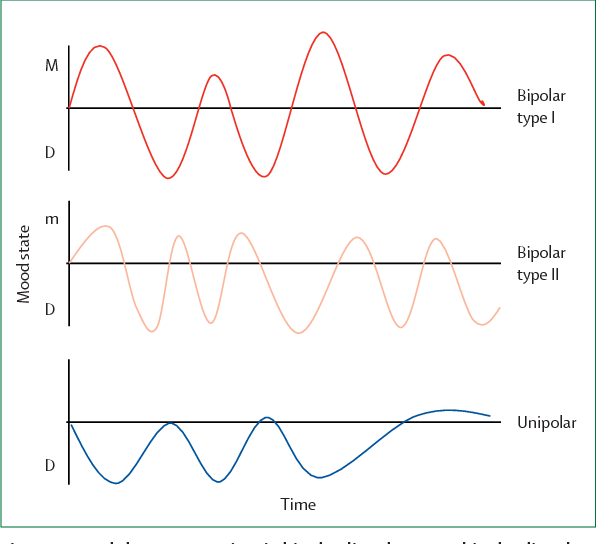
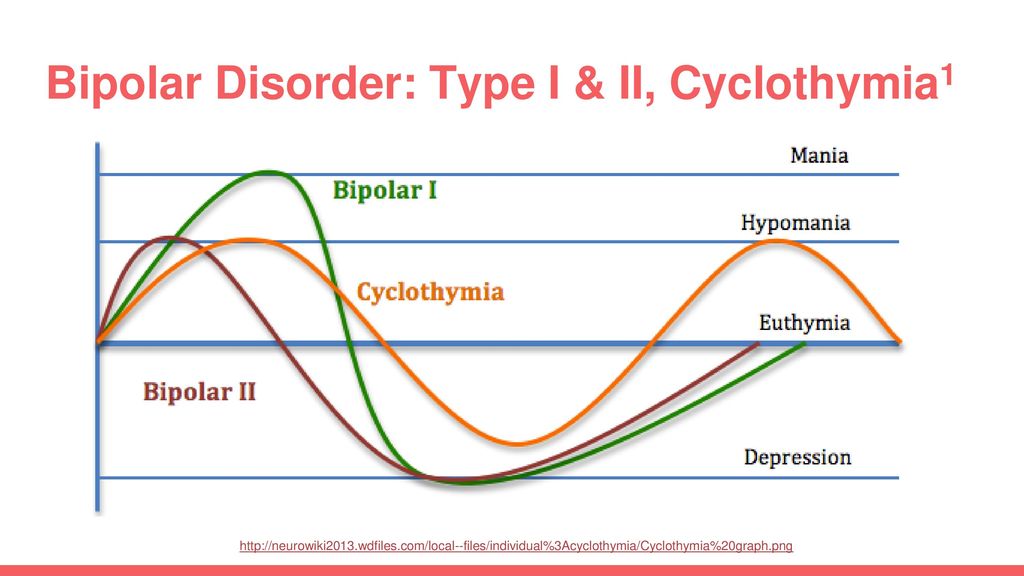
Bipolar Spectrum Disorders (RBS) - cyclothymia, as well as bipolar affective disorder (BAD) of type I and II - polygenic diseases of a multifactorial nature, the development of which is influenced by both genetic factors and and environmental factors. RHD is manifested by disturbances in mood, thinking, and behavior that are characterized by bipolar phases—recurrent episodes of depression and mania in type I bipolar disorder or hypomania in type II bipolar disorder, but mixed episodes are also frequently observed. RHD has traditionally been classified either as part of the spectrum of psychoses or mood disorders (affective disorders). The lifetime prevalence is about 2.1% and does not differ between men (2.2%) and women (2.0%).
According to twin studies, the heritability of bipolar disorder is up to 85%, which indicates a high genetic influence on the risk of developing this disorder and its prognosis. At the same time, it is necessary to take into account environmental factors that also affect the manifestation and course of BAD.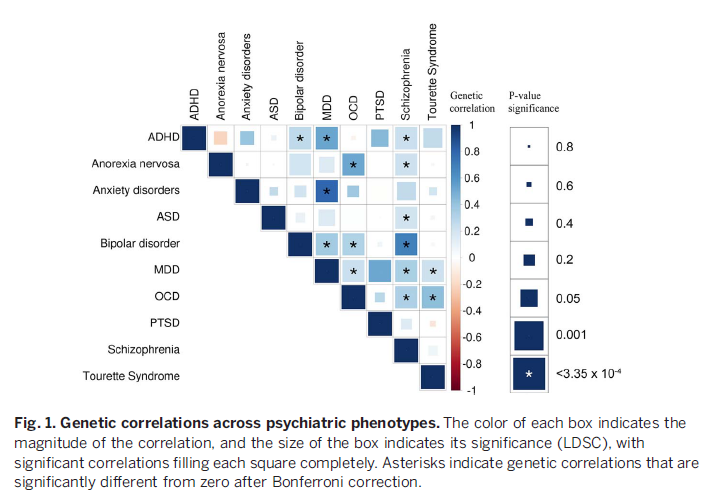 Examples of such factors include perinatal risk factors such as caesarean section, maternal influenza infection, maternal smoking during pregnancy, and late paternal age. Traumatic events in childhood are important, as is the use of psychoactive substances (PS) during adolescence, such as cannabis or other drugs, which can lead to an early onset of bipolar disorder and a more severe course. In addition, various somatic (endocrinological, immune or vascular) diseases can provoke a manifestation of bipolar disorder, as well as the use of drugs (antidepressants, corticosteroids, androgens, isoniazid and chloroquine) and electroconvulsive therapy.
Examples of such factors include perinatal risk factors such as caesarean section, maternal influenza infection, maternal smoking during pregnancy, and late paternal age. Traumatic events in childhood are important, as is the use of psychoactive substances (PS) during adolescence, such as cannabis or other drugs, which can lead to an early onset of bipolar disorder and a more severe course. In addition, various somatic (endocrinological, immune or vascular) diseases can provoke a manifestation of bipolar disorder, as well as the use of drugs (antidepressants, corticosteroids, androgens, isoniazid and chloroquine) and electroconvulsive therapy.
Family studies attempt to answer the question - do disorders tend to aggregate (accumulate) in families? Family design typically compares the incidence of the disorder among first-degree relatives of an affected proband with the incidence of the disorder among relatives of unaffected probands and the general population. The higher frequency of the disorder among relatives of affected probands indicates that the disorder may run in families. However, this does not necessarily mean that only genes are involved in the etiology and pathogenesis of this disorder, as it can occur in families due to common environmental factors (for example, excessive stress, living in an ecologically unfavorable area, etc.).
However, this does not necessarily mean that only genes are involved in the etiology and pathogenesis of this disorder, as it can occur in families due to common environmental factors (for example, excessive stress, living in an ecologically unfavorable area, etc.).
According to family studies, first-degree relatives of affected individuals have a multiple increased risk of bipolar disorder. E.S. Gershon et al. reported a 19-fold increase in the risk of developing bipolar disorder and a 12.4-fold increase in the risk of developing recurrent depressive disorder (RDD). Twin studies (and to a lesser extent, studies with adopted children) have also provided evidence that genes contribute significantly to familial transmission of the disorder. Moreover, bipolar disorder manifesting in early childhood seems to be associated with increased family risk.
As mentioned earlier, although the presence of blood relatives with the same disorder in the family confirms the genetic influence on its development, family studies, unlike twin studies, do not allow us to more accurately quantify the contribution of genetic factors and separate their effects from possible environmental influences or mixed effects. Twin studies comparing groups of mono- and dizygotic pairs of twins can help analyze genetic and environmental contributions. Twin studies typically compare concordance scores for the disorder between monozygotic (MZ) twins (genetically identical) and dizygotic (DZ) twins (sharing half of the genes).
Twin studies comparing groups of mono- and dizygotic pairs of twins can help analyze genetic and environmental contributions. Twin studies typically compare concordance scores for the disorder between monozygotic (MZ) twins (genetically identical) and dizygotic (DZ) twins (sharing half of the genes).
Assuming that the overall environmental influences on MZ twins are no different from environmental influences on DZ twins (equal-conditions assumption), the significantly higher levels of concordance in MZ twins reflect genetic influence itself. However, the level of agreement in MZ twins, which is less than 100%, means that environmental factors do influence the manifestation of a genetic influence - the phenotype of the disease. Twin studies can also be used to assess the contribution of genetic and environmental factors to the variance in susceptibility to the disorder. However, it should be understood that heritability demonstrates the strength of genetic influences for a population, but not for an individual, and estimates of heritability may vary depending on the population under study.
Despite differences in assessment and diagnosis methods, twin studies of bipolar disorder are generally consistent in observing greater agreement between MH twins than DZ twins. This provides strong evidence for the hypothesis that BAD is genetic in nature. Thus, studies have shown that estimates of the heritability of bipolar disorder are in the range of 60-85%, and there is little evidence that the general family environment plays a significant role. However, heritability estimates are less than 100%, indicating the influence of environmental factors
Overall, the available family studies support the findings that: 1) relatives of probands with BD have an increased risk of both BD and RDD; 2) relatives of probands with RDR are at increased risk of RDR, but probably do not have a significantly increased risk of BAD; 3) there is overlap between BAD with other disorders, especially schizophrenia spectrum disorders (In several studies, the risk of BAD among relatives of IAD probands was comparable to or higher than the risk among BAD probands.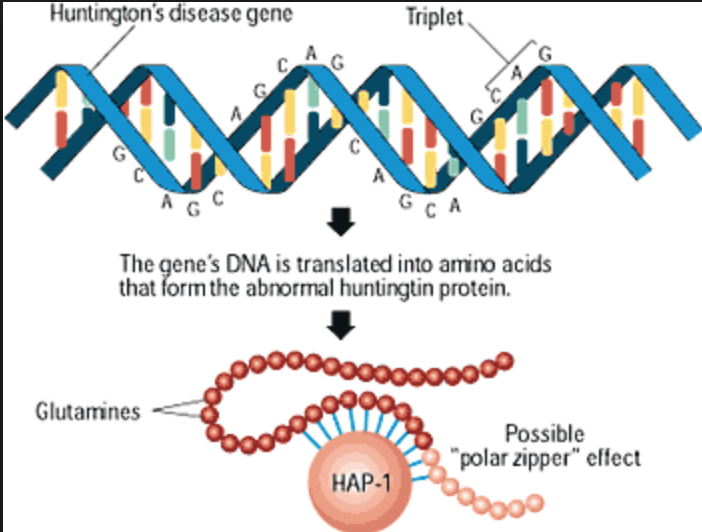 On the other hand, relatives of BAD probands did not a significantly increased risk of RAD was found).
On the other hand, relatives of BAD probands did not a significantly increased risk of RAD was found).
It is also possible that the increased risk of RDR among relatives of probands with bipolar disorder may be overestimated if some relatives with unipolar depression are in fact latent cases of bipolar disorder.
Persons with a burdened history of bipolar disorder have an earlier onset (15 times more often than individuals without a family history of bipolar disorder), a more frequent presence of concomitant psychopathology (panic disorder, psychosis, substance dependence diseases and suicide), and also a more severe course of the underlying disease and a higher risk of hospitalization. In addition, individuals with a family history of bipolar disorder have a higher risk of developing fast cycle bipolar disorder, the most unfavorable subtype of this disorder.
A number of authors suggest that the level and characteristics of the therapeutic response to normothymic drugs, in particular lithium, can serve as a marker for identifying familial subtypes of bipolar disorder. Studies investigating the relationship between familial history of bipolar disorder and lithium response have produced mixed results, but some have found a better response among patients with a familial history, while others have not found a significant or even inverse relationship.
Studies investigating the relationship between familial history of bipolar disorder and lithium response have produced mixed results, but some have found a better response among patients with a familial history, while others have not found a significant or even inverse relationship.
Modern molecular genetic research has achieved some success in studying the nature of BAD. To a greater extent, this is due to the use of the genome-wide association studies method (eng. genome-wide association studies , GWAS). Thus, studies using GWAS, in contrast to studies focused on candidate genes, considered a large number of common variants (single nucleotide polymorphisms) throughout the genome, which increased the chances of detecting genetic associations with BAD. To date, significant genetic associations based on GWAS results have been found in 19chromosomal loci, many of which have been replicated in other studies. BAR-associated genes determine the operation of calcium channels, cell division cycles, cytoskeleton formation, as well as various epigenetic processes, neuronal differentiation, building neuronal connections, regulation of neuronal growth, the operation of the hypothalamic-pituitary-adrenal axis, etc. Several genes, including TRANK1, CACNA1C, ANK3, ITIh4-ITIh5, ZNF804A, and NCAN, have also been associated with schizophrenia.
Several genes, including TRANK1, CACNA1C, ANK3, ITIh4-ITIh5, ZNF804A, and NCAN, have also been associated with schizophrenia.
In addition to the common single nucleotide polymorphism (SNP) genetic polymorphisms found in GWAS, rare genetic variants may also be important for the development of RHD if they have high penetrance (a measure of the phenotypic expression of an allele in a population). ) and therefore may be associated with an increased risk of developing this disorder. Rare variants are classified according to their genomic size into single nucleotide variants, small insertions and deletions, and variants with a large number of copies, including copies of genes or their regions (Copy number variation, CNV; deletions or duplications of large sequences of deoxyribonucleic acid (DNA) in size from 1 thousand to several million base pairs). Thus, some studies have shown the accumulation of rare CNVs in patients with bipolar disorder, especially in patients with an early onset of the disease (from 5 to 18 years). However, these results have not always been replicated in other studies. The latest meta-analysis on this topic showed an association between three CNVs and BAR (duplications at 1q21.1 and 16p11.2 and deletions at 3q29), all of these variants have previously shown an association with schizophrenia.
However, these results have not always been replicated in other studies. The latest meta-analysis on this topic showed an association between three CNVs and BAR (duplications at 1q21.1 and 16p11.2 and deletions at 3q29), all of these variants have previously shown an association with schizophrenia.
However, despite the findings, genetic variants associated with RHD cannot currently be used to predict individual disease risk, its clinical course, or the effectiveness of medical treatment. Thus, the cumulative effect of common alleles with small effects explains only about 25–38% of the phenotypic variance for BAD. In addition, the polygenic nature of these disorders, as well as their heterogeneous phenotype and, as a result, the difficulties in generating truly clinically (phenotypically) homogeneous samples for genetic studies make it unlikely that such a prediction will be possible in the future. It is believed that such studies fail to detect the majority of causal variants that do not reach statistical significance, either due to small effect sizes or low allele frequencies.
A new solution could be an integrated approach using family design and next-generation sequencing methods. Studies with a similar methodology have been able to identify new genetic variants associated with BD that are involved in the development of the cerebral cortex, circadian rhythms, and glutamate neurotransmission processes.
Family-based design is one of the most popular in the field of medical genetic research and has many unique opportunities for studying the risk of developing a particular disease. With its help, it is possible to reduce genetic heterogeneity, and the study of family risks can be very informative for predicting the risk of human disease based on polygenic and common environmental components of genetic risk. Family design provides the maximum probability of detecting specific genetic markers associated not only with the phenotype, but also with potential familial forms of BD. However, despite these strengths, family design is rarely used in current clinical and basic research on psychiatric disorders due to the complexity of data collection and the difficulty in motivating patients and their families to participate in such research.
Obviously, given all the achievements of classical research with family design and the outstanding possibilities of modern genetic technologies, in particular, next generation whole exome sequencing, the most optimal approach may be the complex design of such studies. Each of the methods, taken separately, cannot give the expected result, but the correct combination of these methods provides good chances already at the study planning stage. Family design provides the maximum probability of detecting specific genetic markers associated not only with the nosological phenotype (diagnosis), but also with potential familial and hereditary forms of the disease, and modern genetic technologies make it possible to realize such a probability - to identify reliable and reproducible results.
Author: Kasyanov E.D.
Source: Kasyanov ED, Merculova TV, Kibitov AO, Mazo GE. Genetics of bipolar spectrum disorders: focus on family studies using whole exome sequencing // Genetica [Russian Journal of Genetics].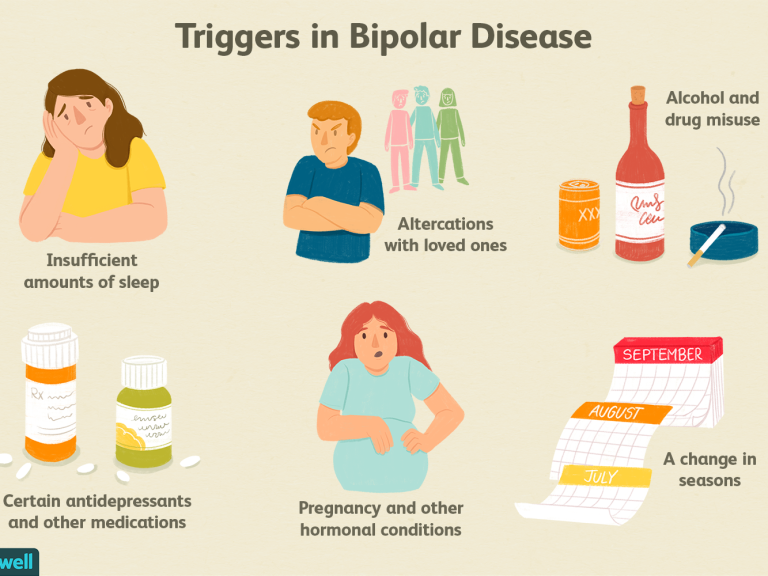 2020. V. 56. No. 7. P. 762-782. doi: 10.31857/S001667582007005X
2020. V. 56. No. 7. P. 762-782. doi: 10.31857/S001667582007005X
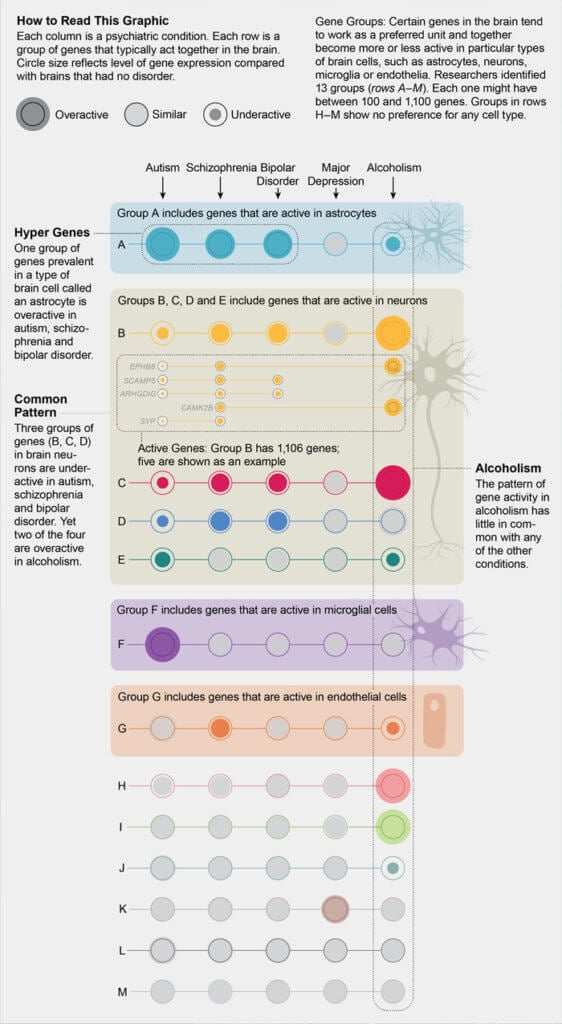
Gene expression profiles 'equate' schizophrenia and bipolar disorder
Researchers analyzed and compared Gene Expression Profiles in the Brain in Five Common Disorders psyche - autism, schizophrenia, depression, bipolar disorder and alcoholism. All diseases, except for alcoholism, turned out to be similar in varying degrees. expression profile, which is confirmed by the presence of common genetic markers. Article published at Science there you can also find an editorial note on the study.
Many mental illnesses have a significant hereditary component, and the same genetic markers often correlate with different diseases at the same time. This may mean that the development for example, schizophrenia, bipolar disorder and depression, goes through a similar mechanism. However, what kind of violations at the molecular level occur in the brain during the development of mental illness, is still unknown.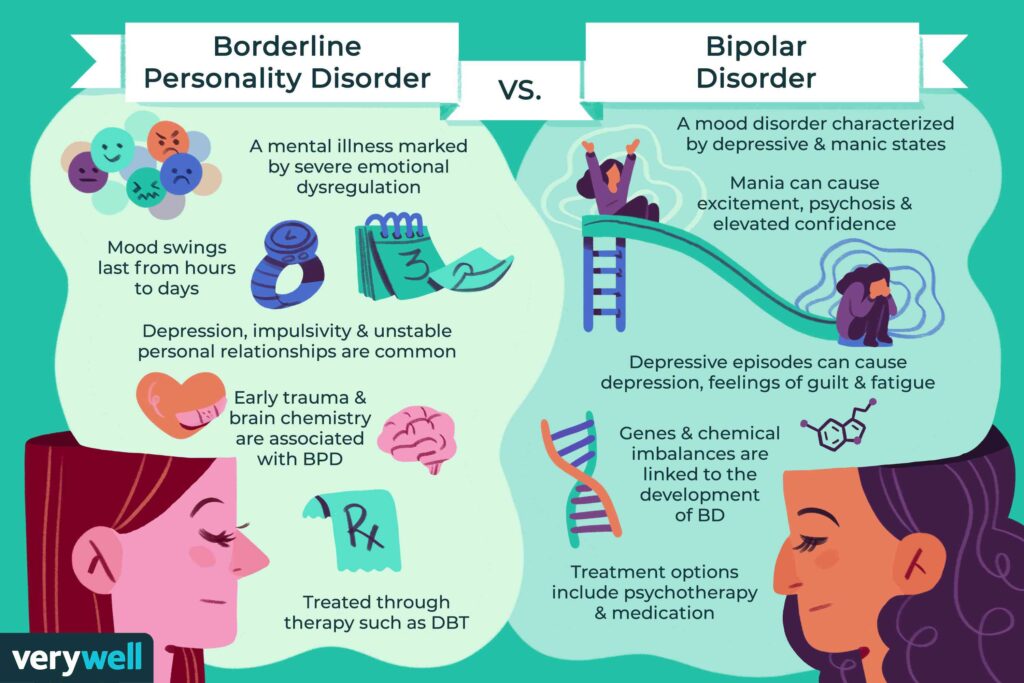 There are no biochemical tests to diagnose patient for a particular disease.
There are no biochemical tests to diagnose patient for a particular disease.
To advance our understanding of the mechanisms mental illness at the molecular level, researchers from the University California (Los Angeles, USA) compared gene expression profiles in the cerebral cortex of 700 a person suffering from the five most common mental diseases. The work was carried out in collaboration with scientific groups commonmind, PsychENCODE and iPSYCH-BROAD, which analyze gene expression in brain tissues taken from patients postmortem.
The sample included 50 former patients with autism, 159 schizophrenia, 94 with bipolar disorder, 87 with major depression disorder and 17 patients with alcoholism. Two were taken as controls. samples - 293 "healthy" samples and 197 samples taken from patients with intestinal inflammation - a disease not associated with the nervous system. The researchers looked at how gene expression profiles patients change compared to controls, and the degree of correlation was determined changes among themselves for different disorders in pairs.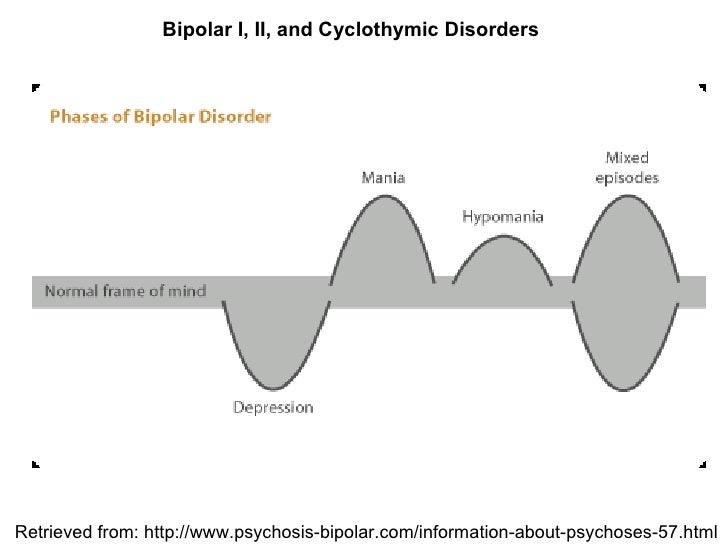
The profiles are most similar to each other expression in schizophrenia and bipolar disorder, diseases that little similar in manifestation, but have common genetic markers. According to experts, this is quite surprising, since the clinical symptoms of bipolar the disorder is usually grouped together with depression (disorders sentiments). However, the expression correlation coefficient for depression and bipolar disorder was three times lower than for schizophrenia and bipolar disorder. In second place in terms of the degree of correlation was the schizophrenia-autism pair. Despite the fact that twin studies have previously indicated the presence of a common hereditary component in depression and alcoholism, the analysis showed that gene expression alcoholism does not correlate with any other disease.
Changes in gene expression also indicated possible molecular mechanisms of disease development. For example, for autism, they found increased activity of genes associated with hyperactivation of microglia - immune cells central nervous system. This led the researchers to speculate that the inflammation is involved in the development of the disease. According to the editor of Science , one of the authors of the study organized a small clinical trial to see how the suppression of microglial activity would affect condition of patients with autism.
This led the researchers to speculate that the inflammation is involved in the development of the disease. According to the editor of Science , one of the authors of the study organized a small clinical trial to see how the suppression of microglial activity would affect condition of patients with autism.
In general, the authors note that the similarity of gene expression profiles between various disorders is consistent with the presence of common genetic markers for these conditions, which, firstly, confirms the significant contribution of genetics to their development, and secondly, indicates the commonality of development mechanisms.
As scientists have previously shown, the development of schizophrenia is probably contributes one of the proteins of the immune system, which is also involved in maturation of synapses - contacts between neurons. About the history of schizophrenia research and how its genetic nature is studied can be read in our materials (1, 2, 3).